By Dror Berel
From a data-science perspective.
You’ve got a fresh new project on your desk, some exciting data, a challenging Kaggle competition, a new client you wish to impress, and you are fully motivated. At first, the problem seems to be well defined, and you even feel comfortable with the task in hand. You have just completed a similar task. This new one should not be much different. Maybe even just a few copy/pastes with some modifications at the edges.
But then it comes… The client / collaborator / boss has just one simple additional request… It usually goes like this:
‘Hmmmm, I wonder how would the results look like if instead of x, we do only a minor change, just do y, or… you know what, let’s try both and see how it affects the results’.
Can the initial tool/solution you chose handle such an adjustment? It may be easy to copy/paste it with a couple of alterations, but what if you have to do it again and again? For how long are you going to stick to your initial plan?
Within the context of machine learning, some examples are:
Tuning ‘let’s see how a different model parameter affects it’
Benchmarking ‘let’s see how various models affect it’
Ensemble ‘let’s try combining the best models together’
Resampling / cross-validation ‘we must inspect for over-fitting’
Imagine adding on top of that some complex, messy, multi-layer, high-throughput genomics data that can easily go into a very fine resolution level (gene expression / mutation / sequence, …),… AND THEN adding multiple layers of various multi-genomic data on top of each other, …AND THEN doing it for multiple cohorts / studies in a meta-analysis level … you may end up with a VERY … BIG … UGLY … MESS!
Sound familiar? Unfortunately, I have been in this situation more than once. As much as I was motivated to please my collaborators, at those times, my tools were…limited, and not sufficient to deliver the broader scope resolution. At that time, I might have not even been aware that a higher level of scope was relevant.

A lot has been written about scope creep in the context of project management. But what would a scientist, who was mostly trained to care about the rightness of the analysis / tools, rather than the ‘management’ of the whole project, have to say about it?
The good news, my friend, is that it is never too late to learn from someone else’s mistakes. Here are couple of lessons, learned the hard way. (No worries, this is not another blog post about reproducible research).
Lesson #1: Begin at the end! Define what your scope is. Do you need to extend it?
Make sure you understand what is the highest expected resolution! Brainstorm what would be the craziest outcomes of your project, and then agree on reasonable expectations within your timeframe and budget.
Have a very detailed, clear, definition of the project scope. For example, is your solution going to handle just one data set, or more? How are you going to validate your results? There are always going to be more methods/data sets for that, but what would be just sufficient enough?
The tricky challenge with scope creep is that the client doesn’t really care or think in terms of “scope”. Their goal is to get a solution that solves a hypothesis, or a business need. Whether their request is within or outside scope is entirely your problem! DEAL WITH IT!
In the context of machine learning, back in the day, I used ad-hoc R packages that do just one multivariate model. They did the work well, but were too specific for the developers domain, and lacked the higher resolution on comparing it with other models, or aggregating other models, or lacking resample implementation. Only later did I learn to utilized machine learning meta/aggregator packages such as mlr, tidymodels (formerly caret), or SuperLearner to extend my scope. Read more about it here.
Lesson #2: Do not reinvent the wheel! There are other experts that know how to do it better than you!
In a role where you are expected to be multidisciplinary, and new tools/methods pop daily that are accessible for everyone to use, it may be a slippery fall into a very deep rabbit hole to explore any new approach. And guess what, nobody want you to waste their time/money on that.
How to bet on the right tool? Ask yourself, what do the experts in that domain use? How mature is the tool they developed? Is it going to be maintained, or deprecated? They of course had their own learning curve, and over time, have perfected their tools to overcome the common pitfalls you are about to discover.
For me, with genomics data, it was Bioconductor Object-oriented S4 classes. Read more here about why that was the best tool for my need. Sure, it wasn’t trivial to learn, but I felt comfortable betting on it when I saw how it is implemented at top academic and industry organizations. I also knew that it was not another open source resource that might die. Instead, it as a government and academia-funded project, powered by the best experts of the domain, open, and free, for all of us to use.
Lesson #3: Found a gap? Be creative, but keep it simple!
But what if something in the analytical pipeline is still not in place? A missing link, nowhere to be found, that would have better fit to the specific need you have, bridging the gap?
Here you might need to get some dirty work done, and stop depending on others to provide you the solution. Another potentially slippery scope creep rabbit hole? Maybe… if you are not careful enough!
How to avoid it? Very easy: Keep it Simple!
Here is a very simple example. Suppose you have to solve an unsupervised problem. There is definitely more than one way to it. Which one to choose? Is the simplest one, suppose ‘hierarchical clustering’, just be good enough to begin with? Implement it, see how it works with the rest of your analytical components (data, scalability, reproducibility), and later on, after things have worked out well as you planned, relax that simplification into a more complex method. Do it very carefully and gradually.
More examples to follow next.
Lesson #4: Do not be afraid to refactor!
Tired of patching and debugging poorly cohesive and poorly-designed code that someone else, maybe even your boss, has written long time ago, before better tools became available? You ask yourself, GRRRRR, this is such an ugly workaround, why not just simply use that new approach that was designed specifically for this task? (see lesson #2).
Yes, it is risky to begin everything from scratch, and sometimes you may not have the resources to do it, but perhaps it is time for a reality check.
But what if the refactor solution will give us different results from what our collaborators are already counting on? Well, if there was indeed a past error/bug/mistake, it is better to face it and acknowledge it now, before even more damage is done. But also remember lesson #3: If you stick to simple solutions at the core, refactoring them under broader wrapping solution should assist in producing similar results.
Lesson #5: go to lesson #1.
Case studies:
Here are two case studies from my own experience working with multi genomic data. (Could easily expand to other types of data, but perhaps that is a topic for a future post).
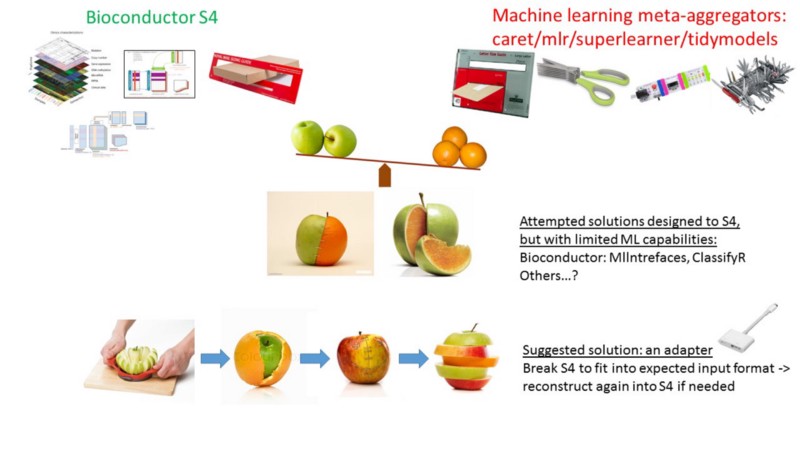
Case Study #1: Bioc2mlr: A utility function to transform Bioconductor’s S4 omic classes into mlr’s task and CPOs.
https://drorberel.github.io/Bioc2mlr/
I love using Bioconductor data containers for genomic data, but I also love machine learning meta-aggregator toolkits for analysis at higher level scope. The only problem was that they were not necessarily compatible with each other.
The S4 object oriented had multiple dimensions (slots), tied in together in complex constraints, that were intentionally designed to meet some purpose. But the machine learning approach was designed for a simplified, flat, two-dimensional, matrix like input structure: columns for the features/variables, and rows for the subjects/observations.
I needed some way of breaking the S4 constrained ties, and flattening it. But unfortunately, to the best of my knowledge, I couldn’t find a way to do so. What should I have done?
Remember lesson #3: Should I spend my time on this task? Well… yes, why not? I felt comfortable enough with both approaches, have already experienced the ins and outs, the soft bellies, and I definitely appreciated the tremendous value of both approaches separately, but also jointly. In fact, creating this adapter package, Bioc2mlr, was not too much effort to do, and if you look at the code itself, you will see relatively simple steps.
Conclusion of case 1: When you have a couple of good tools, but they are not compatible, create a simple new adapter to link them.
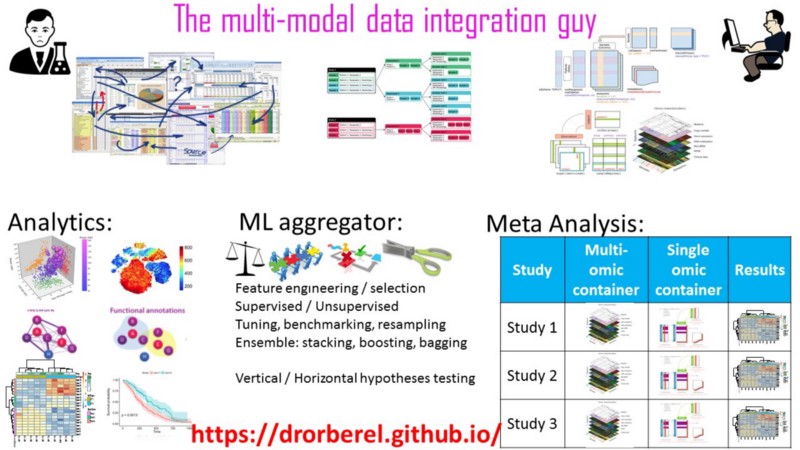
Case study #2: meta analysis
But that wasn’t enough for me…(see lesson #5).
My scope extension required me to provide a solution to even higher level of analysis. Meta-analysis of multiple studies/cohorts, each with a multi-omic data cube, each with a downstream machine learning analytics pipeline, implementing resampling, and all that jazz, across all studies, and at scale. Phewww!
Quite a challenge! How should I address that implementing the above lessons?
Lesson #1: I began at the end. My ‘observation-unit’, row, in a tidy-fashion is not the subject, neither is the gene, nor is just one of the omics. It is the entire study/cohort (that is, a whole data cube) well-compressed into a single object in R. More than one cohort? Not a problem at all. Add as many as rows as you need for more cohorts.
Lesson #2: Didn’t have to invent a new tool. The experts in our field have already figured it out for us. They might have not had this implementation in mind when they did so, but if I can do it, so can you. Just give it a try.
Lesson #3: I found a simple solution. Should I invent/extend a new S4 object oriented class for this type of multi-cohort, multi-omic data? Of course not. There must be a simple solution. My simple solution: a tidy / nested data structure, with non-atomic objects at each cell. Read more about it here.
Lesson #4: Refactor? Well. Maybe I am not there yet now, since so far my (current) scope can handle all my wildest dreams. But if you show me a better approach, perhaps a data.table one (I know), or even in python (god forbid), I would not hesitate to give it a try, even if it is beyond my comfort zone.
Lesson #5: Meta-meta analysis? (Not a typo). Who knows. Maybe one day.
Conclusion of case 2: tidy everything! Even non-atomic objects.
One last piece of advice: Get an expert’s opinion, at least until you become one yourself.
‘If only I had known that before. That could have saved me so much time and effort…’
To the expert, your current challenges are yesterday’s resolution. They had already figured that out when we were still in kindergarten. They have spent their entire career just on that. Shoot them an email, ask a very clear question, with no dependencies proof-of-concepts examples, or case studies to demonstrate your challenge. My experience is that they would be happy to assist if you respect their time and authority.
Final words
When you figure out what type of tool/solution you are passionate about, make it happen! Don’t fool yourself with excuses why it is not a good time for your new tool to be created. Just do it!
Don’t give up. Focus. Decide what you want to achieve. Do not be afraid to extend your scope, but do it with simple solutions! Refactor. It will be worth your time. Maybe not immediately, but in days to come. Be creative!
And last but not least, don’t be shy. Tell everyone about it. Share it with your community. Make the universe a better place with your solution. You may even earn an extra buck on the side. Who knows?
p.s.
This post is dedicated with love to all of my former anxious collaborators / clients / bosses. I appreciate your patience, and wish I would have known the above before. You were there to assist and support me learning these lessons the hard way, for both good and for bad. Let me make it up to you. Shoot me an email and I will redo my old work in just a few lines of code, reflecting my current level of scope.
Check more related topics here: https://drorberel.github.io/
Consultant: currently accepting new projects!
Useful reference:
Clean Coder Blog
_On the Diminished Capacity to Discuss Things Rationally_blog.cleancoder.comScope Creep in Project Management: Definition, Causes & Solutions
_When a project stretches far beyond its original vision, it is called "scope creep". Scope creep in project management…_www.workamajig.com
